A research development that crashed a population of caged mosquitoes could enable elimination of non-target species, says Dr Ricarda Steinbrecher
Gene drive researchers associated with Target Malaria and funded by US DARPA, the GATES Foundation and the UK BBSRC have just managed to crash a population of caged mosquitoes after 7-11 generations.(1)
This is a first, and it has brought this technology beyond the proof of principle. However, in their effort to avoid any resistance to the gene drive, they have focused on a highly stable, ‘conserved’ and vital gene that is found in many species, especially closely related mosquito species. This could enable the gene drive to eliminate populations of multiple non-target species, with very serious implications indeed for biodiversity and ecosystems.
We begin by explaining something about gene drives before focusing on what is different about this latest research just published by Kyrou et al. 2018 in Nature Biotechnology.
Some background to gene drives
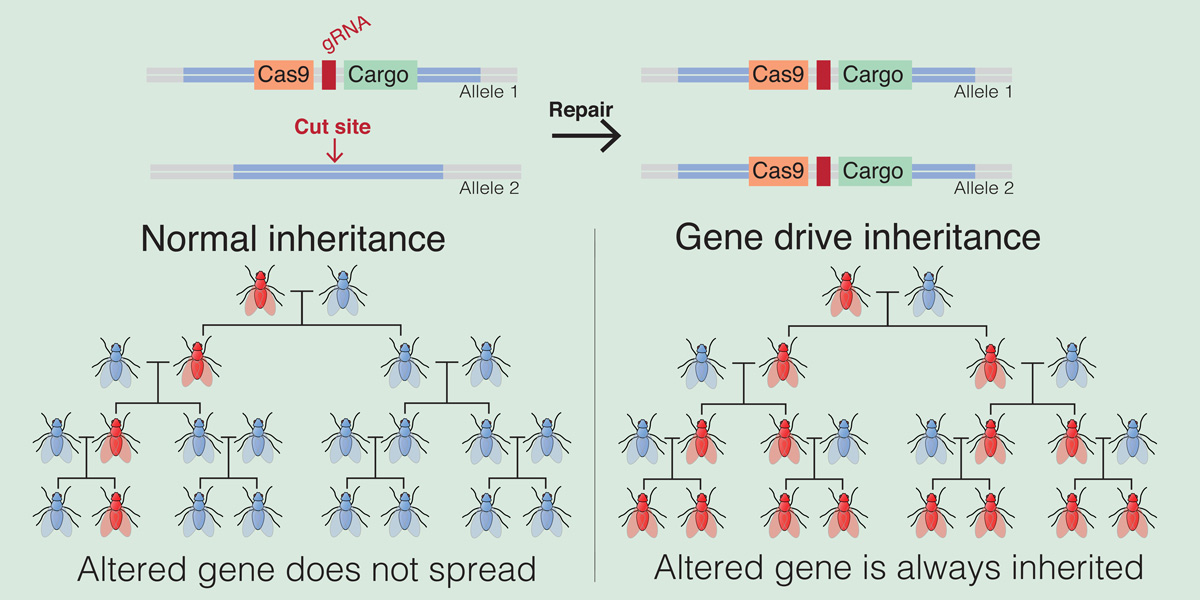
Gene drives are genetic elements that are able to override the rules of inheritance. When genetically engineered and linked to a trait or a handicap they can be used to modify or eradicate a whole population or even an entire species. This idea has largely remained just an idea until quite recently. With the arrival of the genome editing tool CRISPR/Cas9 the field of gene drives and the ability to eliminate undesired species and wild populations got a big boost.
CRISPR gene drives were proposed by Esvelt et al. in 2014,(2) and the proofs of principle came swiftly, published in 2015 and 2016 for fruitflies (Drosophila), two species of mosquitoes and yeast. Gantz & Bier called it a “mutagenic chain reaction” when they delivered the first proof of prinicple (2015) showing that the genome editing tool CRISPR/Cas can – in principle – be turned into a self spreading gene drive capable of altering or eliminating wild populations or potentially whole species.(3) Yet laboratory experiments picked up a flaw in this method – the build-up of ‘resistance’ to CRISPR/Cas, thus stopping the functioning and spread of the gene drive. Why was that?
How the technology works so far
The CRISPR/Cas based gene drives works by recognising and cutting a specific target site in the genome of the organism and copying itself into the gap. If the target site for example is a gene responsible for female fertility, then by cutting it and inserting the gene drive construct containing the CRISPR/Cas information into the target site will disrupt that gene.
Thus CRISPR/Cas is designed and programmed to find a specific target site. If there is for some reason a variation of DNA sequence in the target site, CRISPR/Cas will no longer be able to cut it and insert itself – thus stopping the gene drive from spreading. The altered target site has thus become resistant to the gene drive mechanism.
This might either be because of natural variation of that DNA sequence across the population or it might be that the target sequence had been cut by CRISPR/Cas9, yet the gene drive construct was not inserted, instead the DNA strand was repaired by the ‘wrong’ repair mechanism (the non-homologous end joining repair mechanism), changing the sequence in that process.
If there is an ‘in-frame’ mutation, there may be little consequence to the functioning of a gene, yet the alteration of the sequence will now have created a resistance to the Cas9-based gene drive.(4-7)
New strategy to overcome resistance to gene drives
This has now changed with the new approach published by Kyrou and Hammond et al. (2018).(1) There are two alarming components to this research:
For once, they have for the first time managed to completely crash a caged population, here of Anopheles gambiae mosquitoes, indicating or suggesting they have overcome the resistance problem of CRISPR/Cas9 gene drives in this specific case.
Secondly, they have chosen a highly conserved sex determination gene as the target gene, the doublesex gene. Highly conserved means that the DNA sequence of a gene has remained the same over time on an evloutionary scale and that it has not been changed by random mutations. A highly conserved sequence implies a conserved and highly protected gene, where any alteration to that gene sequence would result in a non-viable life form.
Choosing a highly conserved gene sequence, in particular the sex determination ‘doublesex’ gene, as the gene drive target site means that no viable resistance alleles (gene variants) arise and spread to save the cage population or potentially the wild population. This is a new strategy to overcome gene drive resistance.
Alarming implications
Furthermore, the target sequence is so highly conserved that it can be found across all Anopheles species analysed so far (16 representative Anopheles species from South America, Africa and Asia were analysed by Neafsey and Waterhouse et al. 2015 (8), showing almost complete sequence conservation of the target sequence according to the online methods of Kyrou et al. 2018).
The target sequence is even identical within the Anopheles gambiae species complex, which is comprised of 8 different species, 5 of which were analysed.
This is in fact a serious problem in itself, because once the gene drive complex (or gene drive transgene) crosses into one of the other species, it could potentially run as a gene drive through other populations and species, crashing these populations or eliminating species, with potentially serious ecological and biodiversity consequences.
Such crosses do occur, as shown by introgression studies which identified the movement of genetic elements from one species into the genome of another related species (e.g. Coluzzi et al. 1979 (9); Fontaine et al. 2015 (10). Coluzzi for example reported on 30 different crosses made between 6 of the Anopheles gambiae complex. All the resulting female offspring were fertile, yet only two of the hybrid males, including the cross between gambiae male and quadriannulatus female (A. quadriannulatus is a mosquito feeding on animals).
When studying the reproductive boundaries between Anopheles species there is a degree of fluidity that shows that crosses between different mosquito species do occur. Coluzzi reported an overall 0.1% hybridisation rate in the wild. These findings are highly alarming when looking at the potential consequences of releasing gene drive mosquitoes into the wild, especially when the gene drive target site is so highly conserved as in the current study.
The new strategy thus adds an extra layer of risks and concerns to what was already perceived as a high risk technology.
References
1. Kyrou K, et al. (2018) A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 24 September
2. Esvelt KM, Smidler AL, Catteruccia F, & Church GM (2014) Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3:21.
3. Gantz VM & Bier E (2015) The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science (New York, N.Y.) 348(6233):442-444.
4. Unckless RL, Clark AG, & Messer PW (2017) Evolution of Resistance Against CRISPR/Cas9 Gene Drive. Genetics 205(2):827-841.
5. Champer J, et al. (2017) Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet 13(7):e1006796.
6. Champer J, et al. (2018) Reducing resistance allele formation in CRISPR gene drive. Proceedings of the National Academy of Sciences of the United States of America 115(21):5522-5527.
7. KaramiNejadRanjbar M, et al. (2018) Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proceedings of the National Academy of Sciences of the United States of America 115(24):6189-6194.
8. Neafsey DE, et al. (2015) Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science (New York, N.Y.) 347(6217):9.
9. Coluzzi M, Sabatini A, Petrarca V, & Dideco MA (1979) CHROMOSOMAL DIFFERENTIATION AND ADAPTATION TO HUMAN ENVIRONMENTS IN THE ANOPHELES-GAMBIAE COMPLEX. Trans. Roy. Soc. Trop. Med. Hyg. 73(5):483-497.
10. Fontaine MC, et al. (2015) Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science (New York, N.Y.) 347(6217):7.
Dr Ricarda Steinbrecher is a molecular geneticist and co-director of EcoNexus









