
The row over whether genome-editing techniques should be regulated as genetic modification has given rise to a new terminology of "SDNs". We explain what it means and why we need to know
In July 2018 the European Court of Justice ruled that new forms of genetic engineering, known as genome editing techniques, are just as much genetic modification procedures as the old forms, and that the products of these techniques must be regulated as GMOs. In the wake of that ruling a row has grown up about which types of genome editing and other genetic modification procedures should fall under EU GMO legislation and thus be subjected to safety checks and GMO labelling.
GMWatch has always argued that genome editing techniques (sometimes called GM 2.0) should be regulated at least as strictly as older-style genetic modification techniques using transgenesis – moving genes between organisms or species. In our articles we’ve always used the terms “genome editing” and “genetic modification”.
But there are now whole groups of people who use a new terminology of “SDNs” – short for site-directed nucleases – to distinguish between old and new GM techniques and to argue that the products of certain genome-editing techniques should not be regulated as GMOs. For those who are unfamiliar with SDN terminology, we’ve compiled this short guide to the language and arguments, with the help of molecular geneticist Dr Michael Antoniou.
What exactly are SDNs and why do they matter?
All genetic modifications brought about by genome editing techniques use enzymes called site-directed nucleases (SDNs). A nuclease is an enzyme that is capable of cutting nucleic acid (DNA, RNA). In the case of SDNs they are able to cut across both strands of the DNA double helix bringing about a double-strand break. The “site-directed” nature of SDNs refers to the fact that these enzymes are designed to bring about a DNA double-strand break at a pre-determined location within the genome of the cells or organism being targeted.
The main SDN that is being used to generate gene edited crops is the CRISPR (clustered regularly interspaced short palindromic repeats) system. Other SDNs that can be employed are zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).
The arguments
Companies, researchers, and some scientific and regulatory organizations are arguing that genome editing outcomes known as SDN1 and SDN2 mimic what may happen in nature, do not give rise to GMOs, and should not be regulated as GM procedures. They argue that only large genetic changes, designated as SDN3, which like older-style transgenic techniques involve the addition of whole genes or other DNA elements, give rise to genetically modified organisms that should be assessed for risk.[1]
The language
The enzymes that bring about gene editing-type genetic modifications by cutting both strands of the DNA double helix are called site-directed nucleases or SDN for short. The application of SDNs brings about three possible outcomes, designated as SDN1, SDN2, and SDN3 (Figure 1). All these SDN outcomes start with a targeted double-strand DNA break. However, the events that follow the break are different, depending on the design of the SDN procedure.
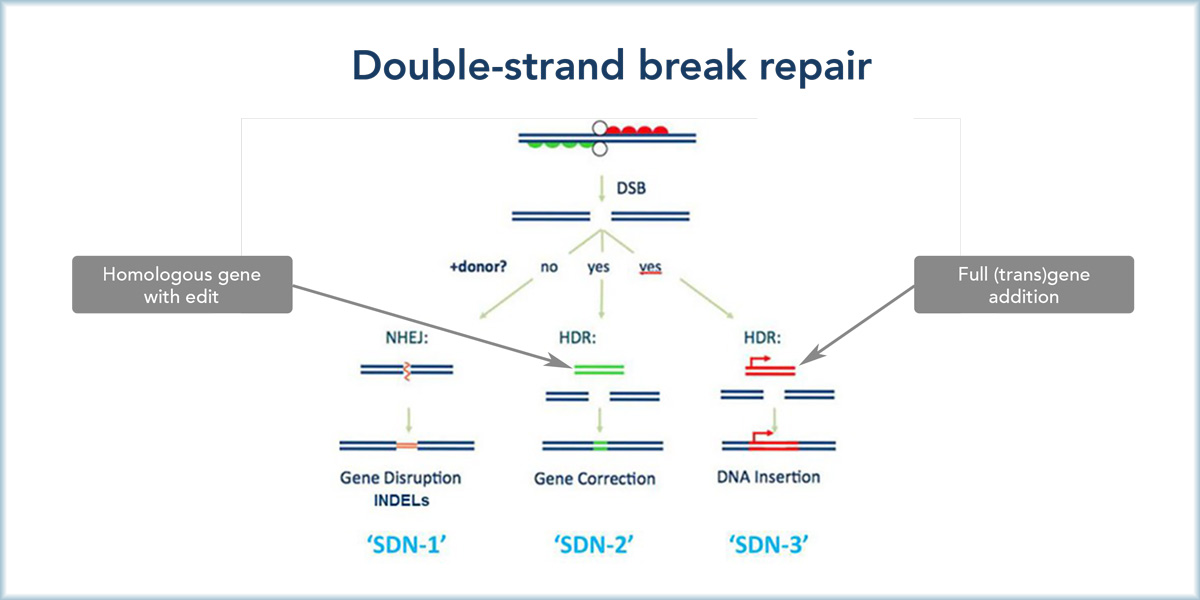
Figure 1. Double-strand break repair in SDN1, SDN2, and SDN3. Definitions: NHEJ (non-homologous end-joining): The cut DNA is rejoined, but while doing this some base pairs may be eaten away or added, resulting in random deletions or additions of nucleotides, or rearrangement of DNA at the cut site. HDR (homology-directed repair): A donor DNA that carries the desired change and is similar in nucleotide sequence to the target site is used to introduce this change at the cut site. In this way you can, in principle, introduce intentional insertions, which can be whole genes, or changes or deletions. The image is adapted from one published by the biotech industry lobby group PRRI.
The three types of SDN outcomes are defined as follows.
SDN1
SDN1 refers to a double-strand break of the DNA helix, which is repaired without the addition of foreign DNA. The repair of the double-strand DNA break is mediated by a process known as non-homologous end joining (NHEJ). The repair mediated by NHEJ results in the re-joining of the broken strands of DNA. However, this re-joining invariably results in either a deletion of DNA base units (nucleotides) or insertion of additional nucleotides at the SDN1-mediated DNA break site. These insertion-deletion events are collectively referred to as “indels”. Thus SDN1 always results in the disruption of the normal nucleotide sequence of the gene being targeted and therefore results in the destruction of its function in part or in whole (technically known as a gene knock-out). (See Figure 1, left side panel).
SDN2
SDN2 also starts with a double-strand break of the DNA helix. However, unlike in the case of SDN1, repair of the DNA break does not take place via NHEJ-mediated indel formation. Instead, a short nucleotide template, which has a very similar nucleotide sequence to the area of the break, is introduced into the cells at the same time as the SDN. The cell then uses this short nucleotide template to repair the DNA break, through a process known as homology-directed repair (HDR). The end result is – in effect – insertion, either in part or in full, of the short nucleotide template at the site of the SDN-mediated double-strand DNA break.
If the DNA repair template differs slightly in its nucleotide sequence from the site of the SDN-induced double-strand break, when it is inserted to repair the break, then a modification in the nucleotide sequence of the gene being targeted will have been brought about (see Figure 1, middle panel).
In this way, genome editors can bring about small but significant alterations in the function of a gene and its products, such as alterations in the activity of an enzyme, conferring new biochemical properties on the organism (e.g., tolerance to a herbicide).
SDN3
SDN3 also initially results in a double-strand break in the DNA. However, similarly to SDN2, at the time when the SDN is introduced into cells, an additional foreign DNA fragment (which is very large compared to the template in SDN2 procedures) is also introduced. This large DNA fragment can contain a complete gene or other genetic material and acts as a template for repair of the double-strand DNA break brought about by the SDN.
As in the case of SDN2, the repair is by insertion of the new genetic material at the site of the double-strand DNA break mediated by the SDN. This takes place as follows. The new genetic material to be inserted is flanked at both ends by extensive nucleotide sequence regions that are the same as (homologous to) those found at the site of the SDN-induced double-strand DNA break. The cell uses these flanking regions of homologous DNA sequence to insert the foreign DNA fragment at the site of the double-strand DNA break, through the HDR process. As a result, a new gene or other genetic material is incorporated into the genome of the organism, imparting upon it novel functions and characteristics (see Figure 1, right side panel).
It is important to note that the final SDN1/2/3 outcomes take place after the SDN has made the DNA double-strand break. The DNA repair takes place via the cell’s natural DNA repair pathways (NHEJ, HDR), which takes place totally independently of the SDN being used. Thus, regardless of how precise is the location of the initial SDN-induced double-strand DNA break, the final SDN1/2/3 outcome is at the mercy of the efficiency and precision of NHEJ and HDR – and there is plenty of evidence that these processes are not precise or predictable.[2,3] Claims of the “precise” nature of gene editing need to be put into this context.
Small changes can have large effects
The use of all types of SDNs and their outcomes (SDN1/2/3) are genetic modification procedures with unintended outcomes, at both the intended on-target and the unintended off-target locations within the genome.[2,3] In the case of crops and foods produced with SDN procedures, such large on-target deletions or rearrangements and/or off-target effects could disrupt the function of multiple genes, which could lead to unexpected toxicity or allergenicity, or undesirable environmental impacts.
On-target or off-target effects can result from modifications that are described as small; that is, where one or a few nucleotides of a gene have been altered. Even changing a single nucleotide within a gene sequence can induce drastic changes in the gene’s function and/or its expression. Such changes can be brought about in SDN1, SDN2, and SDN3 procedures. For example, a change in the function of an enzyme through alteration of its active site can lead to its being able to carry out unintended biochemical reactions.
In addition, genes and their RNA or protein products work in networks. Thus an apparently small change in one gene can affect the function of other members of the network in which the edited gene functions.
Thus even the “small” genetic modifications brought about by genome editing, which are claimed to mimic what may occur in nature and thus not to require regulation, can affect the function of the targeted gene and other gene functions in addition.
For the above reasons, all organisms resulting from SDN1, SDN2, and SDN3 procedures should be subjected to a process-specific GMO-specific risk assessment and regulated as GMOs.
References
1. Sprink T et al (2016). Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts. Plant Cell Rep. 2016; 35: 1493–1506. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4903111/
2. See references collected in: Robinson C, Antoniou M and Fagan J (2018). GMO Myths and Truths, 4th edition. Earth Open Source. https://www.amazon.com/GMO-Myths-Truths-Citizens-Genetically/dp/0993436722/
3. Kosicki M et al (2018). Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nature Biotechnology 36:765–771. https://www.nature.com/articles/nbt.4192
Report: Claire Robinson